Table of Contents
- 1 What is Parallel computation?
- 2 The Global Interpreter Lock
- 3 The Dask delayed function.
- 4 Bag: Parallel Lists for semi-structured data
- 5 Arrays
- 6
dask.arraycontains these algorithms - 7 Performance and Parallelism
- 8 Performance comparision
- 9 Limitations
- 10 Dask DataFrames
- 10.1 When to use
dask.dataframe - 10.2 Create data
- 10.3 Setup
- 10.4 Computations with
dask.dataframe - 10.5 Exercises
- 10.5.1 1.) How many rows are in our dataset?
- 10.5.2 2.) In total, how many non-canceled flights were taken?
- 10.5.3 3.) In total, how many non-cancelled flights were taken from each airport?
- 10.5.4 4.) What was the average departure delay from each airport?
- 10.5.5 5.) What day of the week has the worst average departure delay?
- 10.6 Sharing Intermediate Results
- 10.7 How does this compare to Pandas?
- 10.8 Dask DataFrame Data Model
- 10.9 Converting
CRSDepTimeto a timestamp - 10.10 Exercise: Rewrite above to use a single call to
map_partitions - 10.11 Limitations
- 10.12 Learn More
- 10.1 When to use
- 11 Dask Distributed
- 12 Dask Distributed, Advanced
- 13 Dask Data Storage
Parallelism in Python¶
The last 2 hours of this lecture are modified from the Anaconda Dask tutorial. These lectures are used under licence.
You may find various packages aren’t installed on your system. We recommend you run the following in an Anaconda prompt (on Windows) or a terminal (on Mac) run:
conda env create -f environment.yml
conda activate dask-tutorial
jupyter notebook lecture.ipynb
The first section only uses builtin modules, so we can work through it while that installation completes.
What is Parallel computation?¶
In the simplest sense, parallel computing is the simultaneous use of multiple compute resources to solve a computational problem:
A problem is broken into discrete parts that can be solved concurrently Each part is further broken down to a series of instructions Instructions from each part execute simultaneously on different processors An overall control/coordination mechanism is employed
Blaise Barney, Lawrence Livermore National Laboratory
Threads and Processes¶
To perform a piece of music, one could have a single musician playing many notes at once (for example a pianist, or many musicians playing one note each (for example a string quartet). Similarly, to perform a parallel task on a modern computer, one could have a single instance of a program do many things at once, or have many copies of a program do one thing each. In more exact language, task parallelism on a multi-processor computing system can be multi-threaded or multi-process.
Each version has its own advantages, and which version is best (or whether to combine both!) depends on the nature of the problem to be solved. Threads are lightweight, which make them cheap to create and destroy. They also share resources including program memory, which makes communication between them cheap. However this also requires thought to ensure that two threads don’t modify the same piece of memory at the same time. Processes are larger, and thus more expensive to create temporarily, Communication is also more involved. However, there is no reason processes need to run on the same machine to be able to communicate, so multiprocessing works well at the scale of computer clusters, particularly if the problem being worked on divides naturally into independent parts.
Writing parallel algorithms¶
Not all problems parallelise easily. if each step in a problem depends explictly and nonlinearly on the one before, then it is likely to be hard to divide the workload. Similarly, some problems are “embarassingly parallel”, where a single set of operations needs to be performed on all the elements of an array of data. This is precisely the form of problem which is great to pass to a GPU hardware accelerator.
Syncronous versus Asyncronous computation¶
As soon as a code inteacts with an external process, it can operate in two different modes, a synchrous one, where the call waits each time for the operation to return its result before continuing, or an asyncronous one, in which the original process continues, perhaps checking back later to collect a result. Jobs which absolutely require waiting for them are called “blocking”, while jobs which never need to be waited for are called “non-blocking”. Modern Python 3 contains keywords async, to declare a function as asyncronous, and await to block for a call and collect final results. A useful builtin module, asyncio, gives us some more control over the running of these processes.
import time
import asyncio
def fun1(n, m):
a = [_**2 for _ in range(m)]
time.sleep(n)
return a
async def _fun2a(n):
t = await asyncio.sleep(n)
return t
async def _fun2b(m):
a = [_**2 for _ in range(m)]
return a
async def fun2(n, m):
a = await asyncio.gather(_fun2a(n), _fun2b(m))
return a[0]
t1 = time.time()
a = fun1(2.0, 1000000)
print(time.time()-t1)
t1 = time.time()
a = await fun2(2.0, 1000000)
print(time.time()-t1)
Asyncronous routines are notoriously hard to get straight, and as with many issues to do with parallel programming, can be very hard to debug.
Lazy evalution¶
A concept directly related to asyncronous computation is lazy evaluation. In general, the fastest code is code that does not run. Computer programs frequently contain multiple intermediate values which aren’t necessarily required in a final computation, so if we delay evaluating the value of an expression until it is absolutely need to calculate a later value, we can save both memory and time. This philiosophy is termed “lazy” evaluation, with the opposite standpoint, in which all expressions are evaluated immediately is called “eager” evaluation
from functools import total_ordering
@total_ordering
class Lazy_factorial:
"""Example of a factorial class with lazy evaluation & caching."""
def __init__(self, n):
"""Factorial class, returns n factorial when evaluated."""
self._result = None
self.n = n
def compute(self):
if not self._result:
self._result = self.factorial(self.n)
return self._result
def factorial(self, n):
res = 1
for _ in range(1, n+1):
res *= _
return res
def __repr__(self):
return f'Lazy_factorial({self.n})'
def __lt__(self, other):
return self.n < other.n
def __eq__(self, other):
return self.n == other.n
%time a=Lazy_factorial(1)
%time b=Lazy_factorial(1000)
%time a.compute()
%time b.compute()
%time b.compute()
The threading and multiprocessing modules¶
The builtin threading module provide basic access to allow loops and functions to be multithreaded. Let’s have an example for some simple functions:
import threading
import multiprocessing
import numpy as np
def do_nothing(n):
print(f"Run {n}.")
time.sleep(1)
%%time
threads = []
for i in range(10):
threads.append(threading.Thread(target=do_nothing, args=(i,)))
threads[-1].start()
for thread in threads:
thread.join()
You’ll notice the rather messed up IO. This is because all the different threads are sending their output at the same time. We can fix this by using a lock so that only one thread is allowed to write to screen at a time.
def do_nothing_with_lock(n, lock):
lock.acquire()
print(f"Run {n}.")
lock.release()
time.sleep(1)
%%time
threads = []
lock = threading.Lock()
for i in range(10):
threads.append(threading.Thread(target=do_nothing_with_lock, args=(i, lock)))
threads[-1].start()
for thread in threads:
thread.join()
There are actually a few locks built in to the IO system anyway, which is why the result is as tidy as it is, with the mess confined to new lines only.
The Global Interpreter Lock¶
So far we’ve been speeding up a do nothing function using time.sleep. What happens if we try to do something useful instead? Maybe something in numpy?
arr0 = np.arange(1000000, dtype=float)
arr1 = arr0.copy()
def do_something_in_numpy(arr):
arr[:] = np.sin(arr)
%time do_something_in_numpy(arr1)
arr2 = arr0.copy()
%%time
threads = []
n = 2
N = arr2.size//n
for i in range(n):
threads.append(threading.Thread(target=do_something_in_numpy,
args=(arr2[i*N:(i+1)*N],)))
threads[-1].start()
for thread in threads:
thread.join()
(arr1 == arr2).all()
That worked out ok! Now let’s try some regular Python.
def do_something_in_python(n):
print(f"Run {n}.")
a = [_**2 for _ in range(1000000)]
%%time
# Our serial run
for i in range(10):
do_something_in_python(i)
%%time
threads = []
for i in range(10):
threads.append(threading.Thread(target=do_something_in_python, args=(i,)))
threads[-1].start()
for thread in threads:
thread.join()
Hmm, that doesn’t look so good.
It turns out that for thread safety, and to maximise the speed of serial code, Python implements a single, global lock, which is applied while running most pure Python code. So what’s the solution? The multiprocessing module implements process based parallelism for Python problems, and since these are separate processes, they each have their own GIL.
%%time
processes = []
for i in range(10):
processes.append(multiprocessing.Process(target=do_something_in_python, args=(i,)))
processes[-1].start()
for process in processes:
process.join()
def fn(x):
return x**3
a = range(100)
pool = multiprocessing.Pool(processes=4)
pool.map(fn, a)
You will note we have to write a lot of “boilerplate” code to apply the builtin Python methods for parallel processing. This gets even worse if we are running in a multi-machine computing cluster, so that the processes we wish to communicate with aren’t all running on the same computer. The dask package simplifies a lot of this process. Let’s start a new conda environment supporting the dask tutorial and begin the first Dask notebook.
The Dask delayed function.¶
In this section we parallelize simple for-loop style code with Dask and dask.delayed. Often, this is the only function that you will need to convert functions for use with Dask.
This is a simple way to use dask to parallelize existing codebases or build complex systems. This will also help us to develop an understanding for later sections.
Related Documentation
As we’ll see in the distributed scheduler section, Dask has several ways of executing code in parallel. We’ll use the distributed scheduler by creating a dask.distributed.Client object. For now, this will provide us with some nice diagnostics. We’ll talk about schedulers in depth later.
from dask.distributed import Client
## n_workers could be the number of threads or processes
client = Client(n_workers=4, processes=False)
Basics¶
First let’s make some toy functions, inc and add, that sleep for a while to simulate work. We’ll then time running these functions normally.
In the next section we’ll parallelize this code.
from time import sleep
def inc(x):
sleep(1)
return x + 1
def add(x, y):
sleep(1)
return x + y
%%time
# This takes three seconds to run because we call each
# function sequentially, one after the other
x = inc(1)
y = inc(2)
z = add(x, y)
Parallelize with the dask.delayed decorator¶
Those two increment calls could be called in parallel, because they are totally independent of one-another.
We’ll transform the inc and add functions using the dask.delayed function. When we call the delayed version by passing the arguments, exactly as before, but the original function isn’t actually called yet - which is why the cell execution finishes very quickly.
Instead, a delayed object is made, which keeps track of the function to call and the arguments to pass to it.
from dask import delayed
%%time
# This runs immediately, all it does is build a graph
x = delayed(inc)(1)
y = delayed(inc)(2)
z = delayed(add)(x, y)
This ran immediately, since nothing has really happened yet.
To get the result, call compute. Notice that this runs faster than the original code.
%%time
# This actually runs our computation using a local thread pool
z.compute()
What just happened?¶
The z object is a lazy Delayed object. This object holds everything we need to compute the final result, including references to all of the functions that are required and their inputs and relationship to one-another. We can evaluate the result with .compute() as above or we can visualize the task graph for this value with .visualize().
z
# Look at the task graph for `z`
z.visualize()
Notice that this includes the names of the functions from before, and the logical flow of the outputs of the inc functions to the inputs of add.
Some questions to consider:¶
Why did we go from 3s to 2s? Why weren’t we able to parallelize down to 1s?
What would have happened if the inc and add functions didn’t include the
sleep(1)? Would Dask still be able to speed up this code?What if we have multiple outputs or also want to get access to x or y?
Exercise: Parallelize a for loop¶
for loops are one of the most common things that we want to parallelize. Use dask.delayed on inc and sum to parallelize the computation below:
data = [1, 2, 3, 4, 5, 6, 7, 8]
%%time
# Sequential code
results = []
for x in data:
y = inc(x)
results.append(y)
total = sum(results)
total
%%time
# Your parallel code here. See solutions.ipynb for a model answer.
How do the graph visualizations compare with the given solution, compared to a version with the sum function used directly rather than wrapped with delayed? Can you explain the latter version? You might find the result of the following expression illuminating
delayed(inc)(1) + delayed(inc)(2)
Exercise: Parallelizing a for-loop code with control flow¶
Often we want to delay only some functions, running a few of them immediately. This is especially helpful when those functions are fast and help us to determine what other slower functions we should call. This decision, to delay or not to delay, is usually where we need to be thoughtful when using dask.delayed.
In the example below we iterate through a list of inputs. If that input is even then we want to call inc. If the input is odd then we want to call double. This is_even decision to call inc or double has to be made immediately (not lazily) in order for our graph-building Python code to proceed.
def double(x):
sleep(1)
return 2 * x
def is_even(x):
return not x % 2
data = [1, 2, 3, 4, 5, 6, 7, 8, 9, 10]
%%time
# Sequential code
results = []
for x in data:
if is_even(x):
y = double(x)
else:
y = inc(x)
results.append(y)
total = sum(results)
print(total)
%%time
# Your parallel code here...
# TODO: parallelize the sequential code above using dask.delayed
# You will need to delay some functions, but not all
Some questions to consider:¶
What are other examples of control flow where we can’t use delayed?
What would have happened if we had delayed the evaluation of
is_even(x)in the example above?What are your thoughts on delaying
sum? This function is both computational but also fast to run.
Exercise: Parallelizing a Pandas Groupby Reduction¶
In this exercise we read several CSV files and perform a groupby operation in parallel. We are given sequential code to do this and parallelize it with dask.delayed.
The computation we will parallelize is to compute the mean departure delay per airport from some historical flight data. We will do this by using dask.delayed together with pandas. In a future section we will do this same exercise with dask.dataframe.
Create data¶
Run this code to prep some data.
This downloads and extracts some historical flight data for flights out of NYC between 1990 and 2000. The data is originally from here.
%run prep.py -d flights
Inspect data¶
import os
sorted(os.listdir(os.path.join('data', 'nycflights')))
Read one file with pandas.read_csv and compute mean departure delay¶
import pandas as pd
df = pd.read_csv(os.path.join('data', 'nycflights', '1990.csv'))
df.head()
# What is the schema?
df.dtypes
# What originating airports are in the data?
df.Origin.unique()
# Mean departure delay per-airport for one year
df.groupby('Origin').DepDelay.mean()
Sequential code: Mean Departure Delay Per Airport¶
The above cell computes the mean departure delay per-airport for one year. Here we expand that to all years using a sequential for loop.
from glob import glob
filenames = sorted(glob(os.path.join('data', 'nycflights', '*.csv')))
%%time
sums = []
counts = []
for fn in filenames:
# Read in file
df = pd.read_csv(fn)
# Groupby origin airport
by_origin = df.groupby('Origin')
# Sum of all departure delays by origin
total = by_origin.DepDelay.sum()
# Number of flights by origin
count = by_origin.DepDelay.count()
# Save the intermediates
sums.append(total)
counts.append(count)
# Combine intermediates to get total mean-delay-per-origin
total_delays = sum(sums)
n_flights = sum(counts)
mean = total_delays / n_flights
mean
Parallelize the code above¶
Use dask.delayed to parallelize the code above. Some extra things you will need to know.
Methods and attribute access on delayed objects work automatically, so if you have a delayed object you can perform normal arithmetic, slicing, and method calls on it and it will produce the correct delayed calls.
x = delayed(np.arange)(10) y = (x + 1)[::2].sum() # everything here was delayed
Calling the
.compute()method works well when you have a single output. When you have multiple outputs you might want to use thedask.computefunction:>>> from dask import compute >>> x = delayed(np.arange)(10) >>> y = x ** 2 >>> min_, max_ = compute(y.min(), y.max()) >>> min_, max_ (0, 81)
This way Dask can share the intermediate values (like
y = x**2)
So your goal is to parallelize the code above (which has been copied below) using dask.delayed. You may also want to visualize a bit of the computation to see if you’re doing it correctly.
from dask import compute
%%time
# copied sequential code
sums = []
counts = []
for fn in filenames:
# Read in file
df = pd.read_csv(fn)
# Groupby origin airport
by_origin = df.groupby('Origin')
# Sum of all departure delays by origin
total = by_origin.DepDelay.sum()
# Number of flights by origin
count = by_origin.DepDelay.count()
# Save the intermediates
sums.append(total)
counts.append(count)
# Combine intermediates to get total mean-delay-per-origin
total_delays = sum(sums)
n_flights = sum(counts)
mean = total_delays / n_flights
mean
%%time
# your code here
# ensure the results still match
mean
Some questions to consider:¶
How much speedup did you get? Is this how much speedup you’d expect?
Experiment with where to call
compute. What happens when you call it onsumsandcounts? What happens if you wait and call it onmean?Experiment with delaying the call to
sum. What does the graph look like ifsumis delayed? What does the graph look like if it isn’t?Can you think of any reason why you’d want to do the reduction one way over the other?
Learn More¶
Visit the Delayed documentation. In particular, this delayed screencast will reinforce the concepts you learned here and the delayed best practices document collects advice on using dask.delayed well.
Close the Client¶
Before moving on to the next exercise, make sure to close your client or stop this kernel.
client.close()
Bag: Parallel Lists for semi-structured data¶
Dask-bag excels in processing data that can be represented as a sequence of arbitrary inputs. We’ll refer to this as “messy” data, because it can contain complex nested structures, missing fields, mixtures of data types, etc. The functional programming style fits very nicely with standard Python iteration, such as can be found in the itertools module.
Messy data is often encountered at the beginning of data processing pipelines when large volumes of raw data are first consumed. The initial set of data might be JSON, CSV, XML, or any other format that does not enforce strict structure and datatypes.
For this reason, the initial data massaging and processing is often done with Python lists, dicts, and sets.
These core data structures are optimized for general-purpose storage and processing. Adding streaming computation with iterators/generator expressions or libraries like itertools or toolz let us process large volumes in a small space. If we combine this with parallel processing then we can churn through a fair amount of data.
Dask.bag is a high level Dask collection to automate common workloads of this form. In a nutshell
dask.bag = map, filter, toolz + parallel execution
Related Documentation
Create data¶
%run prep.py -d accounts
Setup¶
Again, we’ll use the distributed scheduler. Schedulers will be explained in depth later.
from dask.distributed import Client
client = Client(n_workers=4)
Creation¶
You can create a Bag from a Python sequence, from files, from data on S3, etc.
We demonstrate using .take() to show elements of the data. (Doing .take(1) results in a tuple with one element)
Note that the data are partitioned into blocks, and there are many items per block. In the first example, the two partitions contain five elements each, and in the following two, each file is partitioned into one or more bytes blocks.
# each element is an integer
import dask.bag as db
b = db.from_sequence([1, 2, 3, 4, 5, 6, 7, 8, 9, 10], npartitions=2)
b.take(3)
# each element is a text file, where each line is a JSON object
# note that the compression is handled automatically
import os
b = db.read_text(os.path.join('data', 'accounts.*.json.gz'))
b.take(1)
# Edit sources.py to configure source locations
import sources
sources.bag_url
# Requires `s3fs` library
# each partition is a remote CSV text file
b = db.read_text(sources.bag_url,
storage_options={'anon': True})
b.take(1)
Manipulation¶
Bag objects hold the standard functional API found in projects like the Python standard library, toolz, or pyspark, including map, filter, groupby, etc..
Operations on Bag objects create new bags. Call the .compute() method to trigger execution, as we saw for Delayed objects.
def is_even(n):
return n % 2 == 0
b = db.from_sequence([1, 2, 3, 4, 5, 6, 7, 8, 9, 10])
c = b.filter(is_even).map(lambda x: x ** 2)
c
# blocking form: wait for completion (which is very fast in this case)
c.compute()
Example: Accounts JSON data¶
We’ve created a fake dataset of gzipped JSON data in your data directory. This is like the example used in the DataFrame example we will see later, except that it has bundled up all of the entires for each individual id into a single record. This is similar to data that you might collect off of a document store database or a web API.
Each line is a JSON encoded dictionary with the following keys
id: Unique identifier of the customer
name: Name of the customer
transactions: List of
transaction-id,amountpairs, one for each transaction for the customer in that file
filename = os.path.join('data', 'accounts.*.json.gz')
lines = db.read_text(filename)
lines.take(3)
Our data comes out of the file as lines of text. Notice that file decompression happened automatically. We can make this data look more reasonable by mapping the json.loads function onto our bag.
import json
js = lines.map(json.loads)
# take: inspect first few elements
js.take(3)
Basic Queries¶
Once we parse our JSON data into proper Python objects (dicts, lists, etc.) we can perform more interesting queries by creating small Python functions to run on our data.
# filter: keep only some elements of the sequence
js.filter(lambda record: record['name'] == 'Alice').take(5)
def count_transactions(d):
return {'name': d['name'], 'count': len(d['transactions'])}
# map: apply a function to each element
(js.filter(lambda record: record['name'] == 'Alice')
.map(count_transactions)
.take(5))
# pluck: select a field, as from a dictionary, element[field]
(js.filter(lambda record: record['name'] == 'Alice')
.map(count_transactions)
.pluck('count')
.take(5))
# Average number of transactions for all of the Alice entries
(js.filter(lambda record: record['name'] == 'Alice')
.map(count_transactions)
.pluck('count')
.mean()
.compute())
Use flatten to de-nest¶
In the example below we see the use of .flatten() to flatten results. We compute the average amount for all transactions for all Alices.
js.filter(lambda record: record['name'] == 'Alice').pluck('transactions').take(3)
(js.filter(lambda record: record['name'] == 'Alice')
.pluck('transactions')
.flatten()
.take(3))
(js.filter(lambda record: record['name'] == 'Alice')
.pluck('transactions')
.flatten()
.pluck('amount')
.take(3))
(js.filter(lambda record: record['name'] == 'Alice')
.pluck('transactions')
.flatten()
.pluck('amount')
.mean()
.compute())
Groupby and Foldby¶
Often we want to group data by some function or key. We can do this either with the .groupby method, which is straightforward but forces a full shuffle of the data (expensive) or with the harder-to-use but faster .foldby method, which does a streaming combined groupby and reduction.
groupby: Shuffles data so that all items with the same key are in the same key-value pairfoldby: Walks through the data accumulating a result per key
Note: the full groupby is particularly bad. In actual workloads you would do well to use foldby or switch to DataFrames if possible.
groupby¶
Groupby collects items in your collection so that all items with the same value under some function are collected together into a key-value pair.
b = db.from_sequence(['Alice', 'Bob', 'Charlie', 'Dan', 'Edith', 'Frank'])
b.groupby(len).compute() # names grouped by length
b = db.from_sequence(list(range(10)))
b.groupby(lambda x: x % 2).compute()
b.groupby(lambda x: x % 2).starmap(lambda k, v: (k, max(v))).compute()
foldby¶
Foldby can be quite odd at first. It is similar to the following functions from other libraries:
When using foldby you provide
A key function on which to group elements
A binary operator such as you would pass to
reducethat you use to perform reduction per each groupA combine binary operator that can combine the results of two
reducecalls on different parts of your dataset.
Your reduction must be associative. It will happen in parallel in each of the partitions of your dataset. Then all of these intermediate results will be combined by the combine binary operator.
is_even = lambda x: x % 2
b.foldby(is_even, binop=max, combine=max).compute()
Example with account data¶
We find the number of people with the same name.
%%time
# Warning, this one takes a while...
result = js.groupby(lambda item: item['name']).starmap(lambda k, v: (k, len(v))).compute()
print(sorted(result))
%%time
# This one is comparatively fast and produces the same result.
from operator import add
def incr(tot, _):
return tot+1
result = js.foldby(key='name',
binop=incr,
initial=0,
combine=add,
combine_initial=0).compute()
print(sorted(result))
Exercise: compute total amount per name¶
We want to groupby (or foldby) the name key, then add up the all of the amounts for each name.
Steps
Create a small function that, given a dictionary like
{'name': 'Alice', 'transactions': [{'amount': 1, 'id': 123}, {'amount': 2, 'id': 456}]}produces the sum of the amounts, e.g.
3Slightly change the binary operator of the
foldbyexample above so that the binary operator doesn’t count the number of entries, but instead accumulates the sum of the amounts.
# Your code here...
DataFrames¶
For the same reasons that Pandas is often faster than pure Python, dask.dataframe can be faster than dask.bag. We will work more with DataFrames later, but from for the bag point of view, they are frequently the end-point of the “messy” part of data ingestion—once the data can be made into a data-frame, then complex split-apply-combine logic will become much more straight-forward and efficient.
You can transform a bag with a simple tuple or flat dictionary structure into a dask.dataframe with the to_dataframe method.
df1 = js.to_dataframe()
df1.head()
This now looks like a well-defined DataFrame, and we can apply Pandas-like computations to it efficiently.
Using a Dask DataFrame, how long does it take to do our prior computation of numbers of people with the same name? It turns out that dask.dataframe.groupby() beats dask.bag.groupby() more than an order of magnitude; but it still cannot match dask.bag.foldby() for this case.
%time df1.groupby('name').id.count().compute().head()
Denormalization¶
This DataFrame format is less-than-optimal because the transactions column is filled with nested data so Pandas has to revert to object dtype, which is quite slow in Pandas. Ideally we want to transform to a dataframe only after we have flattened our data so that each record is a single int, string, float, etc..
def denormalize(record):
# returns a list for every nested item, each transaction of each person
return [{'id': record['id'],
'name': record['name'],
'amount': transaction['amount'],
'transaction-id': transaction['transaction-id']}
for transaction in record['transactions']]
transactions = js.map(denormalize).flatten()
transactions.take(3)
df = transactions.to_dataframe()
df.head()
%%time
# number of transactions per name
# note that the time here includes the data load and ingestion
df.groupby('name')['transaction-id'].count().compute()
Limitations¶
Bags provide very general computation (any Python function.) This generality comes at cost. Bags have the following known limitations
Bag operations tend to be slower than array/dataframe computations in the same way that Python tends to be slower than NumPy/Pandas
Bag.groupbyis slow. You should try to useBag.foldbyif possible. UsingBag.foldbyrequires more thought. Even better, consider creating a normalised dataframe.
Learn More¶
Shutdown¶
client.shutdown()
Arrays¶
 Dask array provides a parallel, larger-than-memory, n-dimensional array using blocked algorithms. Simply put: distributed Numpy.
Dask array provides a parallel, larger-than-memory, n-dimensional array using blocked algorithms. Simply put: distributed Numpy.
Parallel: Uses all of the cores on your computer
Larger-than-memory: Lets you work on datasets that are larger than your available memory by breaking up your array into many small pieces, operating on those pieces in an order that minimizes the memory footprint of your computation, and effectively streaming data from disk.
Blocked Algorithms: Perform large computations by performing many smaller computations
In this notebook, we’ll build some understanding by implementing some blocked algorithms from scratch. We’ll then use Dask Array to analyze large datasets, in parallel, using a familiar NumPy-like API.
Related Documentation
Create data¶
%run prep.py -d random
Setup¶
from dask.distributed import Client
client = Client(n_workers=4)
Blocked Algorithms¶
A blocked algorithm executes on a large dataset by breaking it up into many small blocks.
For example, consider taking the sum of a billion numbers. We might instead break up the array into 1,000 chunks, each of size 1,000,000, take the sum of each chunk, and then take the sum of the intermediate sums.
We achieve the intended result (one sum on one billion numbers) by performing many smaller results (one thousand sums on one million numbers each, followed by another sum of a thousand numbers.)
We do exactly this with Python and NumPy in the following example:
# Load data with h5py
# this creates a pointer to the data, but does not actually load
import h5py
import os
f = h5py.File(os.path.join('data', 'random.hdf5'), mode='r')
dset = f['/x']
Compute sum using blocked algorithm
Before using dask, lets consider the concept of blocked algorithms. We can compute the sum of a large number of elements by loading them chunk-by-chunk, and keeping a running total.
Here we compute the sum of this large array on disk by
Computing the sum of each 1,000,000 sized chunk of the array
Computing the sum of the 1,000 intermediate sums
Note that this is a sequential process in the notebook kernel, both the loading and summing.
# Compute sum of large array, one million numbers at a time
sums = []
for i in range(0, 1000000000, 1000000):
chunk = dset[i: i + 1000000] # pull out numpy array
sums.append(chunk.sum())
total = sum(sums)
print(total)
Exercise: Compute the mean using a blocked algorithm¶
Now that we’ve seen the simple example above try doing a slightly more complicated problem, compute the mean of the array, assuming for a moment that we don’t happen to already know how many elements are in the data. You can do this by changing the code above with the following alterations:
Compute the sum of each block
Compute the length of each block
Compute the sum of the 1,000 intermediate sums and the sum of the 1,000 intermediate lengths and divide one by the other
This approach is overkill for our case but does nicely generalize if we don’t know the size of the array or individual blocks beforehand.
# Compute the mean of the array. A model answer is in solutions.ipynb
dask.array contains these algorithms¶
Dask.array is a NumPy-like library that does these kinds of tricks to operate on large datasets that don’t fit into memory. It extends beyond the linear problems discussed above to full N-Dimensional algorithms and a decent subset of the NumPy interface.
Create dask.array object
You can create a dask.array Array object with the da.from_array function. This function accepts
data: Any object that supports NumPy slicing, likedsetchunks: A chunk size to tell us how to block up our array, like(1000000,)
import dask.array as da
x = da.from_array(dset, chunks=(1000000,))
x
** Manipulate dask.array object as you would a numpy array**
Now that we have an Array we perform standard numpy-style computations like arithmetic, mathematics, slicing, reductions, etc..
The interface is familiar, but the actual work is different. dask_array.sum() does not do the same thing as numpy_array.sum().
What’s the difference?
dask_array.sum() builds an expression of the computation. It does not do the computation yet. numpy_array.sum() computes the sum immediately.
Why the difference?
Dask arrays are split into chunks. Each chunk must have computations run on that chunk explicitly. If the desired answer comes from a small slice of the entire dataset, running the computation over all data would be wasteful of CPU and memory.
result = x.sum()
result
Compute result
Dask.array objects are lazily evaluated. Operations like .sum build up a graph of blocked tasks to execute.
We ask for the final result with a call to .compute(). This triggers the actual computation.
result.compute()
Exercise: Compute the mean¶
And the variance, std, etc.. This should be a small change to the example above.
Look at what other operations you can do with the Jupyter notebook’s tab-completion.
Does this match your result from before?
Performance and Parallelism¶

In our first examples we used for loops to walk through the array one block at a time. For simple operations like sum this is optimal. However for complex operations we may want to traverse through the array differently. In particular we may want the following:
Use multiple cores in parallel
Chain operations on a single blocks before moving on to the next one
Dask.array translates your array operations into a graph of inter-related tasks with data dependencies between them. Dask then executes this graph in parallel with multiple threads. We’ll discuss more about this in the next section.
Example¶
Construct a 20000x20000 array of normally distributed random values broken up into 1000x1000 sized chunks
Take the mean along one axis
Take every 100th element
import numpy as np
import dask.array as da
x = da.random.normal(10, 0.1, size=(20000, 20000), # 400 million element array
chunks=(1000, 1000)) # Cut into 1000x1000 sized chunks
y = x.mean(axis=0)[::100] # Perform NumPy-style operations
x.nbytes / 1e9 # Gigabytes of the input processed lazily
%%time
y.compute() # Time to compute the result
Performance comparision¶
The following experiment was performed on a heavy personal laptop. Your performance may vary. If you attempt the NumPy version then please ensure that you have more than 4GB of main memory.
NumPy: 19s, Needs gigabytes of memory
import numpy as np
%%time
x = np.random.normal(10, 0.1, size=(20000, 20000))
y = x.mean(axis=0)[::100]
y
CPU times: user 19.6 s, sys: 160 ms, total: 19.8 s
Wall time: 19.7 s
Dask Array: 4s, Needs megabytes of memory
import dask.array as da
%%time
x = da.random.normal(10, 0.1, size=(20000, 20000), chunks=(1000, 1000))
y = x.mean(axis=0)[::100]
y.compute()
CPU times: user 29.4 s, sys: 1.07 s, total: 30.5 s
Wall time: 4.01 s
Discussion
Notice that the Dask array computation ran in 4 seconds, but used 29.4 seconds of user CPU time. The numpy computation ran in 19.7 seconds and used 19.6 seconds of user CPU time.
Dask finished faster, but used more total CPU time because Dask was able to transparently parallelize the computation because of the chunk size.
Questions
What happens if the dask chunks=(20000,20000)?
Will the computation run in 4 seconds?
How much memory will be used?
What happens if the dask chunks=(25,25)?
What happens to CPU and memory?
Exercise: Meteorological data¶
There is 2GB of somewhat artifical weather data in HDF5 files in data/weather-big/*.hdf5. We’ll use the h5py library to interact with this data and dask.array to compute on it.
Our goal is to visualize the average temperature on the surface of the Earth for this month. This will require a mean over all of this data. We’ll do this in the following steps
Create
h5py.Datasetobjects for each of the days of data on disk (dsets)Wrap these with
da.from_arraycallsStack these datasets along time with a call to
da.stackCompute the mean along the newly stacked time axis with the
.mean()methodVisualize the result with
matplotlib.pyplot.imshow
%run prep.py -d weather
import h5py
from glob import glob
import os
filenames = sorted(glob(os.path.join('data', 'weather-big', '*.hdf5')))
dsets = [h5py.File(filename, mode='r')['/t2m'] for filename in filenames]
dsets[0]
dsets[0][:5, :5] # Slicing into h5py.Dataset object gives a numpy array
%matplotlib inline
import matplotlib.pyplot as plt
fig = plt.figure(figsize=(16, 8))
plt.imshow(dsets[0][::4, ::4], cmap='RdBu_r');
Integrate with dask.array
Make a list of dask.array objects out of your list of h5py.Dataset objects using the da.from_array function with a chunk size of (500, 500).
arrays = [da.from_array(dset, chunks=(500, 500)) for dset in dsets]
arrays
Stack this list of dask.array objects into a single dask.array object with da.stack
Stack these along the first axis so that the shape of the resulting array is (31, 5760, 11520).
x = da.stack(arrays, axis=0)
x
Plot the mean of this array along the time (0th) axis
# complete the following:
fig = plt.figure(figsize=(16, 8))
plt.imshow(..., cmap='RdBu_r')
Plot the difference of the first day from the mean
# Write your code here.
Exercise: Subsample and store¶
In the above exercise the result of our computation is small, so we can call compute safely. Sometimes our result is still too large to fit into memory and we want to save it to disk. In these cases you can use one of the following two functions
da.store: Store dask.array into any object that supports numpy setitem syntax, e.g.f = h5py.File('myfile.hdf5') output = f.create_dataset(shape=..., dtype=...) da.store(my_dask_array, output)da.to_hdf5: A specialized function that creates and stores adask.arrayobject into anHDF5file.da.to_hdf5('data/myfile.hdf5', '/output', my_dask_array)
The task in this exercise is to use numpy step slicing to subsample the full dataset by a factor of two in both the latitude and longitude direction and then store this result to disk using one of the functions listed above.
As a reminder, Python slicing takes three elements
start:stop:step
>>> L = [1, 2, 3, 4, 5, 6, 7]
>>> L[::3]
[1, 4, 7]
# ...
Example: Lennard-Jones potential¶
The Lennard-Jones is used in partical simuluations in physics, chemistry and engineering. It is highly parallelizable.
First, we’ll run and profile the Numpy version on 7,000 particles.
import numpy as np
# make a random collection of particles
def make_cluster(natoms, radius=40, seed=1981):
np.random.seed(seed)
cluster = np.random.normal(0, radius, (natoms,3))-0.5
return cluster
def lj(r2):
sr6 = (1./r2)**3
pot = 4.*(sr6*sr6 - sr6)
return pot
# build the matrix of distances
def distances(cluster):
diff = cluster[:, np.newaxis, :] - cluster[np.newaxis, :, :]
mat = (diff*diff).sum(-1)
return mat
# the lj function is evaluated over the upper traingle
# after removing distances near zero
def potential(cluster):
d2 = distances(cluster)
dtri = np.triu(d2)
energy = lj(dtri[dtri > 1e-6]).sum()
return energy
cluster = make_cluster(int(7e3), radius=500)
%time potential(cluster)
Notice that the most time consuming function is distances:
# this would open in another browser tab
# %load_ext snakeviz
# %snakeviz potential(cluster)
# alternative simple version given text results in this tab
%prun -s tottime potential(cluster)
Dask version¶
Here’s the Dask version. Only the potential function needs to be rewritten to best utilize Dask.
Note that da.nansum has been used over the full \(NxN\) distance matrix to improve parallel efficiency.
import dask.array as da
# compute the potential on the entire
# matrix of distances and ignore division by zero
def potential_dask(cluster):
d2 = distances(cluster)
energy = da.nansum(lj(d2))/2.
return energy
Let’s convert the NumPy array to a Dask array. Since the entire NumPy array fits in memory it is more computationally efficient to chunk the array by number of CPU cores.
from os import cpu_count
dcluster = da.from_array(cluster, chunks=cluster.shape[0]//cpu_count())
This step should scale quite well with number of cores. The warnings are complaining about dividing by zero, which is why we used da.nansum in potential_dask.
e = potential_dask(dcluster)
%time e.compute()
Limitations¶
Dask Array does not implement the entire numpy interface. Users expecting this will be disappointed. Notably Dask Array has the following failings:
Dask does not implement all of
np.linalg. This has been done by a number of excellent BLAS/LAPACK implementations and is the focus of numerous ongoing academic research projects.Dask Array does not support some operations where the resulting shape depends on the values of the array. For those that it does support (for example, masking one Dask Array with another boolean mask), the chunk sizes will be unknown, which may cause issues with other operations that need to know the chunk sizes.
Dask Array does not attempt operations like
sortwhich are notoriously difficult to do in parallel and are of somewhat diminished value on very large data (you rarely actually need a full sort). Often we include parallel-friendly alternatives liketopk.Dask development is driven by immediate need, and so many lesser used functions, like
np.sometruehave not been implemented purely out of laziness. These would make excellent community contributions.
client.shutdown()
Dask DataFrames¶
We finished the Dask Delayed Chapter by building a parallel dataframe computation over a directory of CSV files using dask.delayed. In this section we use dask.dataframe to automatically build similiar computations, for the common case of tabular computations. Dask dataframes look and feel like Pandas dataframes but they run on the same infrastructure that powers dask.delayed.
In this notebook we use the same airline data as before, but now rather than write for-loops we let dask.dataframe construct our computations for us. The dask.dataframe.read_csv function can take a globstring like "data/nycflights/*.csv" and build parallel computations on all of our data at once.
When to use dask.dataframe¶
Pandas is great for tabular datasets that fit in memory. Dask becomes useful when the dataset you want to analyze is larger than your machine’s RAM. The demo dataset we’re working with is only about 200MB, so that you can download it in a reasonable time, but dask.dataframe will scale to datasets much larger than memory.

The dask.dataframe module implements a blocked parallel DataFrame object that mimics a large subset of the Pandas DataFrame. One Dask DataFrame is comprised of many in-memory pandas DataFrames separated along the index. One operation on a Dask DataFrame triggers many pandas operations on the constituent pandas DataFrames in a way that is mindful of potential parallelism and memory constraints.
Related Documentation
Main Take-aways
Dask DataFrame should be familiar to Pandas users
The partitioning of dataframes is important for efficient execution
Create data¶
%run prep.py -d flights
Setup¶
from dask.distributed import Client
client = Client(n_workers=4)
We create artifical data.
from prep import accounts_csvs
accounts_csvs()
import os
import dask
filename = os.path.join('data', 'accounts.*.csv')
filename
Filename includes a glob pattern *, so all files in the path matching that pattern will be read into the same Dask DataFrame.
import dask.dataframe as dd
df = dd.read_csv(filename)
df.head()
# load and count number of rows
len(df)
What happened here?
Dask investigated the input path and found that there are three matching files
a set of jobs was intelligently created for each chunk - one per original CSV file in this case
each file was loaded into a pandas dataframe, had
len()applied to itthe subtotals were combined to give you the final grand total.
Real Data¶
Lets try this with an extract of flights in the USA across several years. This data is specific to flights out of the three airports in the New York City area.
df = dd.read_csv(os.path.join('data', 'nycflights', '*.csv'),
parse_dates={'Date': [0, 1, 2]})
Notice that the respresentation of the dataframe object contains no data - Dask has just done enough to read the start of the first file, and infer the column names and dtypes.
df
We can view the start and end of the data
df.head()
df.tail() # this fails
What just happened?¶
Unlike pandas.read_csv which reads in the entire file before inferring datatypes, dask.dataframe.read_csv only reads in a sample from the beginning of the file (or first file if using a glob). These inferred datatypes are then enforced when reading all partitions.
In this case, the datatypes inferred in the sample are incorrect. The first n rows have no value for CRSElapsedTime (which pandas infers as a float), and later on turn out to be strings (object dtype). Note that Dask gives an informative error message about the mismatch. When this happens you have a few options:
Specify dtypes directly using the
dtypekeyword. This is the recommended solution, as it’s the least error prone (better to be explicit than implicit) and also the most performant.Increase the size of the
samplekeyword (in bytes)Use
assume_missingto makedaskassume that columns inferred to beint(which don’t allow missing values) are actually floats (which do allow missing values). In our particular case this doesn’t apply.
In our case we’ll use the first option and directly specify the dtypes of the offending columns.
df = dd.read_csv(os.path.join('data', 'nycflights', '*.csv'),
parse_dates={'Date': [0, 1, 2]},
dtype={'TailNum': str,
'CRSElapsedTime': float,
'Cancelled': bool})
df.tail() # now works
Computations with dask.dataframe¶
We compute the maximum of the DepDelay column. With just pandas, we would loop over each file to find the individual maximums, then find the final maximum over all the individual maximums
maxes = []
for fn in filenames:
df = pd.read_csv(fn)
maxes.append(df.DepDelay.max())
final_max = max(maxes)
We could wrap that pd.read_csv with dask.delayed so that it runs in parallel. Regardless, we’re still having to think about loops, intermediate results (one per file) and the final reduction (max of the intermediate maxes). This is just noise around the real task, which pandas solves with
df = pd.read_csv(filename, dtype=dtype)
df.DepDelay.max()
dask.dataframe lets us write pandas-like code, that operates on larger than memory datasets in parallel.
%time df.DepDelay.max().compute()
This writes the delayed computation for us and then runs it.
Some things to note:
As with
dask.delayed, we need to call.compute()when we’re done. Up until this point everything is lazy.Dask will delete intermediate results (like the full pandas dataframe for each file) as soon as possible.
This lets us handle datasets that are larger than memory
This means that repeated computations will have to load all of the data in each time (run the code above again, is it faster or slower than you would expect?)
As with Delayed objects, you can view the underlying task graph using the .visualize method:
# notice the parallelism
df.DepDelay.max().visualize()
Exercises¶
In this section we do a few dask.dataframe computations. If you are comfortable with Pandas then these should be familiar. You will have to think about when to call compute.
1.) How many rows are in our dataset?¶
If you aren’t familiar with pandas, how would you check how many records are in a list of tuples?
# Your code here
2.) In total, how many non-canceled flights were taken?¶
With pandas, you would use boolean indexing.
# Your code here
3.) In total, how many non-cancelled flights were taken from each airport?¶
Hint: use df.groupby.
# Your code here
4.) What was the average departure delay from each airport?¶
Note, this is the same computation you did in the previous notebook (is this approach faster or slower?)
# Your code here
5.) What day of the week has the worst average departure delay?¶
# Your code here
Sharing Intermediate Results¶
When computing all of the above, we sometimes did the same operation more than once. For most operations, dask.dataframe hashes the arguments, allowing duplicate computations to be shared, and only computed once.
For example, lets compute the mean and standard deviation for departure delay of all non-canceled flights. Since dask operations are lazy, those values aren’t the final results yet. They’re just the recipe required to get the result.
If we compute them with two calls to compute, there is no sharing of intermediate computations.
non_cancelled = df[~df.Cancelled]
mean_delay = non_cancelled.DepDelay.mean()
std_delay = non_cancelled.DepDelay.std()
%%time
mean_delay_res = mean_delay.compute()
std_delay_res = std_delay.compute()
But lets try by passing both to a single compute call.
%%time
mean_delay_res, std_delay_res = dask.compute(mean_delay, std_delay)
Using dask.compute takes roughly 1/2 the time. This is because the task graphs for both results are merged when calling dask.compute, allowing shared operations to only be done once instead of twice. In particular, using dask.compute only does the following once:
the calls to
read_csvthe filter (
df[~df.Cancelled])some of the necessary reductions (
sum,count)
To see what the merged task graphs between multiple results look like (and what’s shared), you can use the dask.visualize function (we might want to use filename='graph.pdf' to zoom in on the graph better):
dask.visualize(mean_delay, std_delay)
How does this compare to Pandas?¶
Pandas is more mature and fully featured than dask.dataframe. If your data fits in memory then you should use Pandas. The dask.dataframe module gives you a limited pandas experience when you operate on datasets that don’t fit comfortably in memory.
During this tutorial we provide a small dataset consisting of a few CSV files. This dataset is 45MB on disk that expands to about 400MB in memory. This dataset is small enough that you would normally use Pandas.
We’ve chosen this size so that exercises finish quickly. Dask.dataframe only really becomes meaningful for problems significantly larger than this, when Pandas breaks with the dreaded
MemoryError: ...
Furthermore, the distributed scheduler allows the same dataframe expressions to be executed across a cluster. To enable massive “big data” processing, one could execute data ingestion functions such as read_csv, where the data is held on storage accessible to every worker node (e.g., amazon’s S3), and because most operations begin by selecting only some columns, transforming and filtering the data, only relatively small amounts of data need to be communicated between the machines.
Dask.dataframe operations use pandas operations internally. Generally they run at about the same speed except in the following two cases:
Dask introduces a bit of overhead, around 1ms per task. This is usually negligible.
When Pandas releases the GIL (coming to
groupbyin the next version)dask.dataframecan call several pandas operations in parallel within a process, increasing speed somewhat proportional to the number of cores. For operations which don’t release the GIL, multiple processes would be needed to get the same speedup.
Dask DataFrame Data Model¶
For the most part, a Dask DataFrame feels like a pandas DataFrame. So far, the biggest difference we’ve seen is that Dask operations are lazy; they build up a task graph instead of executing immediately (more details coming in Schedulers). This lets Dask do operations in parallel and out of core.
In Dask Arrays, we saw that a dask.array was composed of many NumPy arrays, chunked along one or more dimensions.
It’s similar for dask.dataframe: a Dask DataFrame is composed of many pandas DataFrames. For dask.dataframe the chunking happens only along the index.
We call each chunk a partition, and the upper / lower bounds are divisions. Dask can store information about the divisions. For now, partitions come up when you write custom functions to apply to Dask DataFrames
Converting CRSDepTime to a timestamp¶
This dataset stores timestamps as HHMM, which are read in as integers in read_csv:
crs_dep_time = df.CRSDepTime.head(10)
crs_dep_time
To convert these to timestamps of scheduled departure time, we need to convert these integers into pd.Timedelta objects, and then combine them with the Date column.
In pandas we’d do this using the pd.to_timedelta function, and a bit of arithmetic:
import pandas as pd
# Get the first 10 dates to complement our `crs_dep_time`
date = df.Date.head(10)
# Get hours as an integer, convert to a timedelta
hours = crs_dep_time // 100
hours_timedelta = pd.to_timedelta(hours, unit='h')
# Get minutes as an integer, convert to a timedelta
minutes = crs_dep_time % 100
minutes_timedelta = pd.to_timedelta(minutes, unit='m')
# Apply the timedeltas to offset the dates by the departure time
departure_timestamp = date + hours_timedelta + minutes_timedelta
departure_timestamp
Custom code and Dask Dataframe¶
We could swap out pd.to_timedelta for dd.to_timedelta and do the same operations on the entire dask DataFrame. But let’s say that Dask hadn’t implemented a dd.to_timedelta that works on Dask DataFrames. What would you do then?
dask.dataframe provides a few methods to make applying custom functions to Dask DataFrames easier:
Here we’ll just be discussing map_partitions, which we can use to implement to_timedelta on our own:
# Look at the docs for `map_partitions`
help(df.CRSDepTime.map_partitions)
The basic idea is to apply a function that operates on a DataFrame to each partition.
In this case, we’ll apply pd.to_timedelta.
hours = df.CRSDepTime // 100
# hours_timedelta = pd.to_timedelta(hours, unit='h')
hours_timedelta = hours.map_partitions(pd.to_timedelta, unit='h')
minutes = df.CRSDepTime % 100
# minutes_timedelta = pd.to_timedelta(minutes, unit='m')
minutes_timedelta = minutes.map_partitions(pd.to_timedelta, unit='m')
departure_timestamp = df.Date + hours_timedelta + minutes_timedelta
departure_timestamp
departure_timestamp.head()
Exercise: Rewrite above to use a single call to map_partitions¶
This will be slightly more efficient than two separate calls, as it reduces the number of tasks in the graph.
def compute_departure_timestamp(df):
pass # TODO: implement this
departure_timestamp = df.map_partitions(compute_departure_timestamp)
departure_timestamp.head()
Limitations¶
What doesn’t work?¶
Dask.dataframe only covers a small but well-used portion of the Pandas API. This limitation is for two reasons:
The Pandas API is huge
Some operations are genuinely hard to do in parallel (e.g. sort)
Additionally, some important operations like set_index work, but are slower
than in Pandas because they include substantial shuffling of data, and may write out to disk.
Learn More¶
client.shutdown()
Dask Distributed¶
As we have seen so far, Dask allows you to simply construct graphs of tasks with dependencies, as well as have graphs created automatically for you using functional, Numpy or Pandas syntax on data collections. None of this would be very useful, if there weren’t also a way to execute these graphs, in a parallel and memory-aware way. So far we have been calling thing.compute() or dask.compute(thing) without worrying what this entails. Now we will discuss the options available for that execution, and in particular, the distributed scheduler, which comes with additional functionality.
Dask comes with four available schedulers:
“threaded”: a scheduler backed by a thread pool
“processes”: a scheduler backed by a process pool
“single-threaded” (aka “sync”): a synchronous scheduler, good for debugging
distributed: a distributed scheduler for executing graphs on multiple machines, see below.
To select one of these for computation, you can specify at the time of asking for a result, e.g.,
myvalue.compute(scheduler="single-threaded") # for debugging
or set the current default, either temporarily or globally
with dask.config.set(scheduler='processes'):
# set temporarily for this block only
myvalue.compute()
dask.config.set(scheduler='processes')
# set until further notice
Lets see the difference for the familiar case of the flights data
%run prep.py -d flights
import dask.dataframe as dd
import os
df = dd.read_csv(os.path.join('data', 'nycflights', '*.csv'),
parse_dates={'Date': [0, 1, 2]},
dtype={'TailNum': object,
'CRSElapsedTime': float,
'Cancelled': bool})
# Maximum average non-cancelled delay grouped by Airport
largest_delay = df[~df.Cancelled].groupby('Origin').DepDelay.mean().max()
largest_delay
# each of the following gives the same results (you can check!)
# any surprises?
import time
for sch in ['threading', 'processes', 'sync']:
t0 = time.time()
r = largest_delay.compute(scheduler=sch)
t1 = time.time()
print(f"{sch:>10}, {t1 - t0:0.4f} s; result, {r:0.2f} hours")
Some Questions to Consider:¶
How much speedup is possible for this task (hint, look at the graph).
Given how many cores are on this machine, how much faster could the parallel schedulers be than the single-threaded scheduler.
How much faster was using threads over a single thread? Why does this differ from the optimal speedup?
Why is the multiprocessing scheduler so much slower here?
The threaded scheduler is a fine choice for working with large datasets out-of-core on a single machine, as long as the functions being used release the GIL most of the time. NumPy and pandas release the GIL in most places, so the threaded scheduler is the default for dask.array and dask.dataframe. The distributed scheduler, perhaps with processes=False, will also work well for these workloads on a single machine.
For workloads that do hold the GIL, as is common with dask.bag and custom code wrapped with dask.delayed, we recommend using the distributed scheduler, even on a single machine. Generally speaking, it’s more intelligent and provides better diagnostics than the processes scheduler.
https://docs.dask.org/en/latest/scheduling.html provides some additional details on choosing a scheduler.
For scaling out work across a cluster, the distributed scheduler is required.
Making a cluster¶
Simple method¶
The dask.distributed system is composed of a single centralized scheduler and one or more worker processes. Deploying a remote Dask cluster involves some additional effort. But doing things locally is just involves creating a Client object, which lets you interact with the “cluster” (local threads or processes on your machine). For more information see here.
Note that Client() takes a lot of optional arguments, to configure the number of processes/threads, memory limits and other
from dask.distributed import Client
# Setup a local cluster.
# By default this sets up 1 worker per core
client = Client()
client.cluster
Be sure to click the Dashboard link to open up the diagnostics dashboard.
Executing with the distributed client¶
Consider some trivial calculation, such as we’ve used before, where we have added sleep statements in order to simulate real work being done.
from dask import delayed
import time
def inc(x):
time.sleep(5)
return x + 1
def dec(x):
time.sleep(3)
return x - 1
def add(x, y):
time.sleep(7)
return x + y
By default, creating a Client makes it the default scheduler. Any calls to .compute will use the cluster your client is attached to, unless you specify otherwise, as above.
x = delayed(inc)(1)
y = delayed(dec)(2)
total = delayed(add)(x, y)
total.compute()
The tasks will appear in the web UI as they are processed by the cluster and, eventually, a result will be printed as output of the cell above. Note that the kernel is blocked while waiting for the result. The resulting tasks block graph might look something like below. Hovering over each block gives which function it related to, and how long it took to execute. 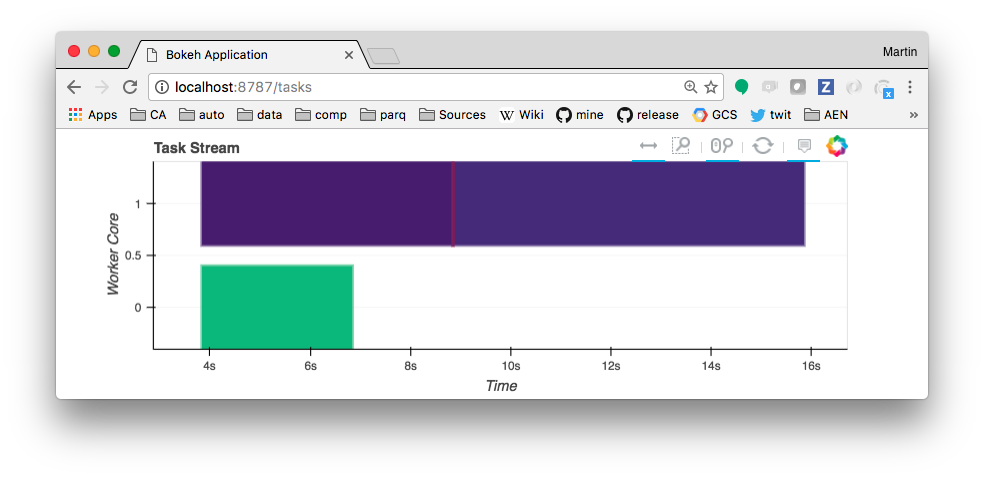
You can also see a simplified version of the graph being executed on Graph pane of the dashboard, so long as the calculation is in-flight.
Let’s return to the flights computation from before, and see what happens on the dashboard (you may wish to have both the notebook and dashboard side-by-side). How did does this perform compared to before?
%time largest_delay.compute()
In this particular case, this should be as fast or faster than the best case, threading, above. Why do you suppose this is? You should start your reading here, and in particular note that the distributed scheduler was a complete rewrite with more intelligence around sharing of intermediate results and which tasks run on which worker. This will result in better performance in some cases, but still larger latency and overhead compared to the threaded scheduler, so there will be rare cases where it performs worse. Fortunately, the dashboard now gives us a lot more diagnostic information. Look at the Profile page of the dashboard to find out what takes the biggest fraction of CPU time for the computation we just performed?
If all you want to do is execute computations created using delayed, or run calculations based on the higher-level data collections, then that is about all you need to know to scale your work up to cluster scale. However, there is more detail to know about the distributed scheduler that will help with efficient usage. See the chapter Distributed, Advanced.
Exercise¶
Run the following computations while looking at the diagnostics page. In each case what is taking the most time?
# Number of flights
_ = len(df)
# Number of non-cancelled flights
_ = len(df[~df.Cancelled])
# Number of non-cancelled flights per-airport
_ = df[~df.Cancelled].groupby('Origin').Origin.count().compute()
# Average departure delay from each airport?
_ = df[~df.Cancelled].groupby('Origin').DepDelay.mean().compute()
# Average departure delay per day-of-week
_ = df.groupby(df.Date.dt.dayofweek).DepDelay.mean().compute()
client.shutdown()
Dask Distributed, Advanced¶
Distributed futures¶
from dask.distributed import Client
c = Client(n_workers=4)
c.cluster
In chapter Distributed, we showed that executing a calculation (created using delayed) with the distributed executor is identical to any other executor. However, we now have access to additional functionality, and control over what data is held in memory.
To begin, the futures interface (derived from the built-in concurrent.futures) allow map-reduce like functionality. We can submit individual functions for evaluation with one set of inputs, or evaluated over a sequence of inputs with submit() and map(). Notice that the call returns immediately, giving one or more futures, whose status begins as “pending” and later becomes “finished”. There is no blocking of the local Python session.
Here is the simplest example of submit in action:
def inc(x):
return x + 1
fut = c.submit(inc, 1)
fut
We can re-execute the following cell as often as we want as a way to poll the status of the future. This could of course be done in a loop, pausing for a short time on each iteration. We could continue with our work, or view a progressbar of work still going on, or force a wait until the future is ready.
In the meantime, the status dashboard (link above next to the Cluster widget) has gained a new element in the task stream, indicating that inc() has completed, and the progress section at the problem shows one task complete and held in memory.
fut
Possible alternatives you could investigate:
from dask.distributed import wait, progress
progress(fut)
would show a progress bar in this notebook, rather than having to go to the dashboard. This progress bar is also asynchronous, and doesn’t block the execution of other code in the meanwhile.
wait(fut)
would block and force the notebook to wait until the computation pointed to by fut was done. However, note that the result of inc() is sitting in the cluster, it would take no time to execute the computation now, because Dask notices that we are asking for the result of a computation it already knows about. More on this later.
# grab the information back - this blocks if fut is not ready
c.gather(fut)
# equivalent action when only considering a single future
# fut.result()
Here we see an alternative way to execute work on the cluster: when you submit or map with the inputs as futures, the computation moves to the data rather than the other way around, and the client, in the local Python session, need never see the intermediate values. This is similar to building the graph using delayed, and indeed, delayed can be used in conjunction with futures. Here we use the delayed object total from before.
# Some trivial work that takes time
# repeated from the Distributed chapter.
from dask import delayed
import time
def inc(x):
time.sleep(5)
return x + 1
def dec(x):
time.sleep(3)
return x - 1
def add(x, y):
time.sleep(7)
return x + y
x = delayed(inc)(1)
y = delayed(dec)(2)
total = delayed(add)(x, y)
# notice the difference from total.compute()
# notice that this cell completes immediately
fut = c.compute(total)
fut
c.gather(fut) # waits until result is ready
Client.submit¶
submit takes a function and arguments, pushes these to the cluster, returning a Future representing the result to be computed. The function is passed to a worker process for evaluation. Note that this cell returns immediately, while computation may still be ongoing on the cluster.
fut = c.submit(inc, 1)
fut
This looks a lot like doing compute(), above, except now we are passing the function and arguments directly to the cluster. To anyone used to concurrent.futures, this will look familiar. This new fut behaves the same way as the one above. Note that we have now over-written the previous definition of fut, which will get garbage-collected, and, as a result, that previous result is released by the cluster
Exercise: Rebuild the above delayed computation using Client.submit instead¶
The arguments passed to submit can be futures from other submit operations or delayed objects. The former, in particular, demonstrated the concept of moving the computation to the data which is one of the most powerful elements of programming with Dask.
# Your code here
Each futures represents a result held, or being evaluated by the cluster. Thus we can control caching of intermediate values - when a future is no longer referenced, its value is forgotten. In the solution, above, futures are held for each of the function calls. These results would not need to be re-evaluated if we chose to submit more work that needed them.
We can explicitly pass data from our local session into the cluster using scatter(), but usually better is to construct functions that do the loading of data within the workers themselves, so that there is no need to serialise and communicate the data. Most of the loading functions within Dask, sudh as dd.read_csv, work this way. Similarly, we normally don’t want to gather() results that are too big in memory.
The full API of the distributed scheduler gives details of interacting with the cluster, which remember, can be on your local machine or possibly on a massive computational resource.
The futures API offers a work submission style that can easily emulate the map/reduce paradigm (see c.map()) that may be familiar to many people. The intermediate results, represented by futures, can be passed to new tasks without having to bring the pull locally from the cluster, and new work can be assigned to work on the output of previous jobs that haven’t even begun yet.
Generally, any Dask operation that is executed using .compute() can be submitted for asynchronous execution using c.compute() instead, and this applies to all collections. Here is an example with the calculation previously seen in the Bag chapter. We have replaced the .compute() method there with the distributed client version, so, again, we could continue to submit more work (perhaps based on the result of the calculation), or, in the next cell, follow the progress of the computation. A similar progress-bar appears in the monitoring UI page.
%run prep.py -d accounts
import dask.bag as db
import os
import json
filename = os.path.join('data', 'accounts.*.json.gz')
lines = db.read_text(filename)
js = lines.map(json.loads)
f = c.compute(js.filter(lambda record: record['name'] == 'Alice')
.pluck('transactions')
.flatten()
.pluck('amount')
.mean())
from dask.distributed import progress
# note that progress must be the last line of a cell
# in order to show up
progress(f)
# get result.
c.gather(f)
# release values by deleting the futures
del f, fut, x, y, total
Persist¶
Considering which data should be loaded by the workers, as opposed to passed, and which intermediate values to persist in worker memory, will in many cases determine the computation efficiency of a process.
In the example here, we repeat a calculation from the Array chapter - notice that each call to compute() is roughly the same speed, because the loading of the data is included every time.
%run prep.py -d random
import h5py
import os
f = h5py.File(os.path.join('data', 'random.hdf5'), mode='r')
dset = f['/x']
import dask.array as da
x = da.from_array(dset, chunks=(1000000,))
%time x.sum().compute()
%time x.sum().compute()
If, instead, we persist the data to RAM up front (this takes a few seconds to complete - we could wait() on this process), then further computations will be much faster.
# changes x from a set of delayed prescriptions
# to a set of futures pointing to data in RAM
# See this on the UI dashboard.
x = c.persist(x)
%time x.sum().compute()
%time x.sum().compute()
Naturally, persisting every intermediate along the way is a bad idea, because this will tend to fill up all available RAM and make the whole system slow (or break!). The ideal persist point is often at the end of a set of data cleaning steps, when the data is in a form which will get queried often.
Exercise: how is the memory associated with x released, once we know we are done with it?
Asynchronous computation¶

One benefit of using the futures API is that you can have dynamic computations that adjust as things progress. Here we implement a simple naive search by looping through results as they come in, and submit new points to compute as others are still running.
Watching the diagnostics dashboard as this runs you can see computations are being concurrently run while more are being submitted. This flexibility can be useful for parallel algorithms that require some level of synchronization.
Lets perform a very simple minimization using dynamic programming. The function of interest is known as Rosenbrock:
# a simple function with interesting minima
import time
def rosenbrock(point):
"""Compute the rosenbrock function and return the point and result"""
time.sleep(0.1)
score = (1 - point[0])**2 + 2 * (point[1] - point[0]**2)**2
return point, score
Initial setup, including creating a graphical figure. We use Bokeh for this, which allows for dynamic update of the figure as results come in.
from bokeh.io import output_notebook, push_notebook
from bokeh.models.sources import ColumnDataSource
from bokeh.plotting import figure, show
import numpy as np
output_notebook()
# set up plot background
N = 500
x = np.linspace(-5, 5, N)
y = np.linspace(-5, 5, N)
xx, yy = np.meshgrid(x, y)
d = (1 - xx)**2 + 2 * (yy - xx**2)**2
d = np.log(d)
p = figure(x_range=(-5, 5), y_range=(-5, 5))
p.image(image=[d], x=-5, y=-5, dw=10, dh=10, palette="Spectral11");
We start off with a point at (0, 0), and randomly scatter test points around it. Each evaluation takes ~100ms, and as result come in, we test to see if we have a new best point, and choose random points around that new best point, as the search box shrinks.
We print the function value and current best location each time we have a new best value.
from dask.distributed import as_completed
from random import uniform
scale = 5 # Intial random perturbation scale
best_point = (0, 0) # Initial guess
best_score = float('inf') # Best score so far
startx = [uniform(-scale, scale) for _ in range(10)]
starty = [uniform(-scale, scale) for _ in range(10)]
# set up plot
source = ColumnDataSource({'x': startx, 'y': starty, 'c': ['grey'] * 10})
p.circle(source=source, x='x', y='y', color='c')
t = show(p, notebook_handle=True)
# initial 10 random points
futures = [c.submit(rosenbrock, (x, y)) for x, y in zip(startx, starty)]
iterator = as_completed(futures)
for res in iterator:
# take a completed point, is it an improvement?
point, score = res.result()
if score < best_score:
best_score, best_point = score, point
print(score, point)
x, y = best_point
newx, newy = (x + uniform(-scale, scale), y + uniform(-scale, scale))
# update plot
source.stream({'x': [newx], 'y': [newy], 'c': ['grey']}, rollover=20)
push_notebook(document=t)
# add new point, dynamically, to work on the cluster
new_point = c.submit(rosenbrock, (newx, newy))
iterator.add(new_point) # Start tracking new task as well
# Narrow search and consider stopping
scale *= 0.99
if scale < 0.001:
break
point
Debugging¶
When something goes wrong in a distributed job, it is hard to figure out what the problem was and what to do about it. When a task raises an exception, the exception will show up when that result, or other result that depend upon it, is gathered.
Consider the following delayed calculation to be computed by the cluster. As usual, we get back a future, which the cluster is working on to compute (this happens very slowly for the trivial procedure).
@delayed
def ratio(a, b):
return a // b
@delayed
def summation(*a):
return sum(*a)
ina = [5, 25, 30]
inb = [5, 5, 6]
out = summation([ratio(a, b) for (a, b) in zip(ina, inb)])
f = c.compute(out)
f
We only get to know what happened when we gather the result (this is also true for out.compute(), except we could not have done other stuff in the meantime). For the first set of inputs, it works fine.
c.gather(f)
But if we introduce bad input, an exception is raised. The exception happens in ratio, but only comes to our attention when calculating summation.
ina = [5, 25, 30]
inb = [5, 0, 6]
out = summation([ratio(a, b) for (a, b) in zip(ina, inb)])
f = c.compute(out)
c.gather(f)
The display in this case makes the origin of the exception obvious, but this is not always the case. How should this be debugged, how would we go about finding out the exact conditions that caused the exception?
The first step, of course, is to write well-tested code which makes appropriate assertions about its input and clear warnings and error messages when something goes wrong. This applies to all code.
The most typical thing to do is to execute some portion of the computation in the local thread, so that we can run the Python debugger and query the state of things at the time that the exception happened. Obviously, this cannot be performed on the whole data-set when dealing with Big Data on a cluster, but a suitable sample will probably do even then.
import dask
with dask.config.set(scheduler="sync"):
# do NOT use c.compute(out) here - we specifically do not
# want the distributed scheduler
out.compute()
# uncomment to enter post-mortem debugger
# %debug
The trouble with this approach is that Dask is meant for the execution of large datasets/computations - you probably can’t simply run the whole thing
in one local thread, else you wouldn’t have used Dask in the first place. So the code above should only be used on a small part of the data that also exchibits the error.
Furthermore, the method will not work when you are dealing with futures (such as f, above, or after persisting) instead of delayed-based computations.
As alternative, you can ask the scheduler to analyze your calculation and find the specific sub-task responsible for the error, and pull only it and its dependnecies locally for execution.
c.recreate_error_locally(f)
# uncomment to enter post-mortem debugger
# %debug
Finally, there are errors other than exceptions, when we need to look at the state of the scheduler/workers. In the standard “LocalCluster” we started, we have direct access to these.
[(k, v.state) for k, v in c.cluster.scheduler.tasks.items() if v.exception is not None]
Dask Data Storage¶
 Efficient storage can dramatically improve performance, particularly when operating repeatedly from disk.
Efficient storage can dramatically improve performance, particularly when operating repeatedly from disk.
Decompressing text and parsing CSV files is expensive. One of the most effective strategies with medium data is to use a binary storage format like HDF5. Often the performance gains from doing this is sufficient so that you can switch back to using Pandas again instead of using dask.dataframe.
In this section we’ll learn how to efficiently arrange and store your datasets in on-disk binary formats. We’ll use the following:
Pandas
HDFStoreformat on top ofHDF5Categoricals for storing text data numerically
Main Take-aways
Storage formats affect performance by an order of magnitude
Text data will keep even a fast format like HDF5 slow
A combination of binary formats, column storage, and partitioned data turns one second wait times into 80ms wait times.
Create data¶
%run prep.py -d accounts
Read CSV¶
First we read our csv data as before.
CSV and other text-based file formats are the most common storage for data from many sources, because they require minimal pre-processing, can be written line-by-line and are human-readable. Since Pandas’ read_csv is well-optimized, CSVs are a reasonable input, but far from optimized, since reading required extensive text parsing.
import os
filename = os.path.join('data', 'accounts.*.csv')
filename
import dask.dataframe as dd
df_csv = dd.read_csv(filename)
df_csv.head()
Write to HDF5¶
HDF5 and netCDF are binary array formats very commonly used in the scientific realm.
Pandas contains a specialized HDF5 format, HDFStore. The dd.DataFrame.to_hdf method works exactly like the pd.DataFrame.to_hdf method.
target = os.path.join('data', 'accounts.h5')
target
# convert to binary format, takes some time up-front
%time df_csv.to_hdf(target, '/data')
# same data as before
df_hdf = dd.read_hdf(target, '/data')
df_hdf.head()
Compare CSV to HDF5 speeds¶
We do a simple computation that requires reading a column of our dataset and compare performance between CSV files and our newly created HDF5 file. Which do you expect to be faster?
%time df_csv.amount.sum().compute()
%time df_hdf.amount.sum().compute()
Sadly they are about the same, or perhaps even slower.
The culprit here is names column, which is of object dtype and thus hard to store efficiently. There are two problems here:
How do we store text data like
namesefficiently on disk?Why did we have to read the
namescolumn when all we wanted wasamount
1. Store text efficiently with categoricals¶
We can use Pandas categoricals to replace our object dtypes with a numerical representation. This takes a bit more time up front, but results in better performance.
More on categoricals at the pandas docs and this blogpost.
# Categorize data, then store in HDFStore
%time df_hdf.categorize(columns=['names']).to_hdf(target, '/data2')
# It looks the same
df_hdf = dd.read_hdf(target, '/data2')
df_hdf.head()
# But loads more quickly
%time df_hdf.amount.sum().compute()
This is now definitely faster than before. This tells us that it’s not only the file type that we use but also how we represent our variables that influences storage performance.
How does the performance of reading depend on the scheduler we use? You can try this with threaded, processes and distributed.
However this can still be better. We had to read all of the columns (names and amount) in order to compute the sum of one (amount). We’ll improve further on this with parquet, an on-disk column-store. First though we learn about how to set an index in a dask.dataframe.
Exercise¶
fastparquet is a library for interacting with parquet-format files, which are a very common format in the Big Data ecosystem, and used by tools such as Hadoop, Spark and Impala.
target = os.path.join('data', 'accounts.parquet')
df_csv.categorize(columns=['names']).to_parquet(target, storage_options={"has_nulls": True}, engine="fastparquet")
Investigate the file structure in the resultant new directory - what do you suppose those files are for?
to_parquet comes with many options, such as compression, whether to explicitly write NULLs information (not necessary in this case), and how to encode strings. You can experiment with these, to see what effect they have on the file size and the processing times, below.
ls -l data/accounts.parquet/
df_p = dd.read_parquet(target)
# note that column names shows the type of the values - we could
# choose to load as a categorical column or not.
df_p.dtypes
Rerun the sum computation above for this version of the data, and time how long it takes. You may want to try this more than once - it is common for many libraries to do various setup work when called for the first time.
%time df_p.amount.sum().compute()
When archiving data, it is common to sort and partition by a column with unique identifiers, to facilitate fast look-ups later. For this data, that column is id. Time how long it takes to retrieve the rows corresponding to id==100 from the raw CSV, from HDF5 and parquet versions, and finally from a new parquet version written after applying set_index('id').
# df_p.set_index('id').to_parquet(...)
Remote files¶
Dask can access various cloud- and cluster-oriented data storage services such as Amazon S3 or HDFS
Advantages:
scalable, secure storage
Disadvantages:
network speed becomes bottleneck
The way to set up dataframes (and other collections) remains very similar to before. Note that the data here is available anonymously, but in general an extra parameter storage_options= can be passed with further details about how to interact with the remote storage.
taxi = dd.read_csv('s3://nyc-tlc/trip data/yellow_tripdata_2015-*.csv',
storage_options={'anon': True})
Warning: operations over the Internet can take a long time to run. Such operations work really well in a cloud clustered set-up, e.g., amazon EC2 machines reading from S3, Microsoft Azure VMs or Google compute machines reading from GCS.
